Papers
2024
-
 Optimizing Biophysically-Plausible Large-Scale Circuit Models With Deep Neural NetworksTianchu Zeng*, Fang Tian* , Shaoshi Zhang, Gustavo Deco, Theodore D. Satterthwaite, Avram Holmes, and B.T. Thomas Yeo2024
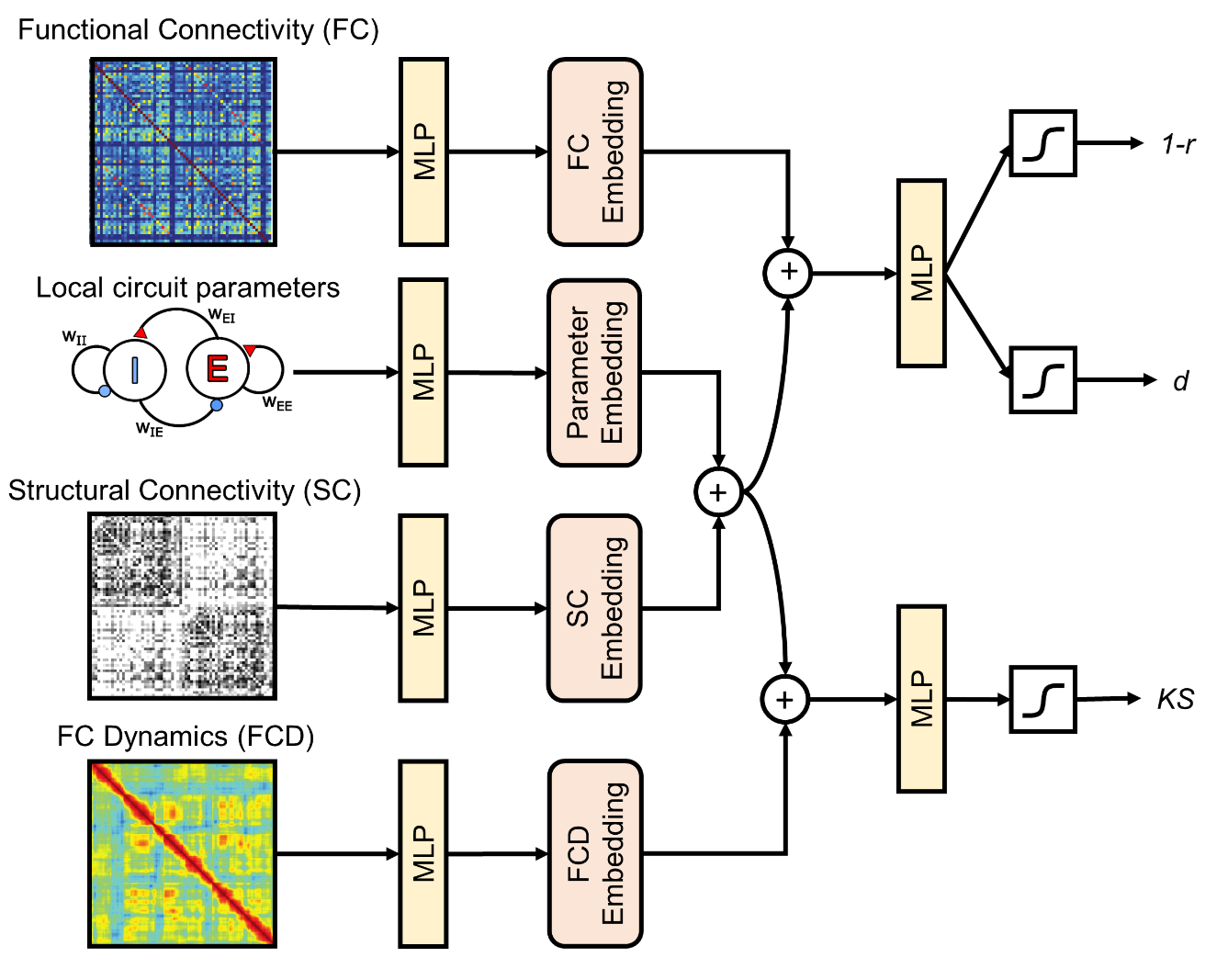
Optimizing Biophysically-Plausible Large-Scale Circuit Models With Deep Neural NetworksTianchu Zeng*, Fang Tian* , Shaoshi Zhang, Gustavo Deco, Theodore D. Satterthwaite, Avram Holmes, and B.T. Thomas Yeo2024Biophysically plausible large-circuit models provide valuable mechanistic insights into neural dynamics. However, setting up a model involves numerous free parameters, which are often manually set, leading to assumptions that may not align with biological plausibility. In contrast, optimizing these parameters results in more biologically plausible models. Previous papers have shown that spatially heterogeneous parameters improve the model performance in generating realistic neural dynamics. Prior works have employed iterative forward simulations via Euler integration to optimize the parameters, which require solving ordinary differential equations (ODE) for all regions simultaneously and each time step sequentially. This process poses a computational hurdle for an efficient model inversion. To tackle this problem, we propose DELSSOME (Deep Learning for Surrogate Statistics Optimization in Mean Field Modeling), which integrates deep learning models into neural mass model framework to circumvent the need to solve ODEs. These deep learning models are designed to learn surrogate statistics, which directly provide outputs required for optimization, from the input local circuit parameters. Our experiments demonstrate that our approach leads to a remarkable 500,000% speed-up of parameter optimization process compared to the traditional approach while achieving comparable performances. Moreover, our approach successfully replicates previous findings on excitation-inhibition ratio changes related to neurodevelopment and cognition. Overall, our results suggest the efficiency and broader applicability of the proposed approach in uncovering intricate neural dynamics.
- Aligning AI models with Editable Explanations (In Progress)2024
2023
-
 Automated ECG Diagnosis using an Explainable AI FrameworkFang Tian , and Brian Y. Lim2023
Automated ECG Diagnosis using an Explainable AI FrameworkFang Tian , and Brian Y. Lim2023Cardiovascular disease (CVD) is a primary cause of mortality globally, and the electrocardiogram (ECG) is a commonly used diagnostic tool for its detection. While Artificial Intelligence (AI) has shown an exceptional predictive ability for CVD, the lack of interpretability has deterred medical professionals from its use. To address this, we developed an explainable AI (XAI) framework that integrates ECG rules expressed in the form of first-order logic (FOL). The framework can uncover the underlying model’s impressions of interpretable ECG features, which can be crucial for cardiologists to understand the diagnosis predictions generated by our system. Our experiments demonstrate the benefits of incorporating ECG rules into ECG AI such as improved performance and the ability to generate a diagnosis report that provides insights into how the model derived the predicted diagnoses. Overall, our XAI framework represents a great step forward in integrating domain knowledge into ECG AI models and enhancing their interpretability.